
Targets for antiviral treatment vary but usually involve an individual viral component, such as a protein or a glycoprotein motif. One of the more intriguing directions over the last decade has been the prevention of virus activity in the cell particularly against RNA viruses. Called RNA interference (RNAi), small, synthetic RNA molecules bind to their viral RNA complements preventing replication and virulence. Studies have shown benefit against a myriad of infections and has also proved to work against cellular RNA in cancer cells.
In the hunt for potential RNAi candidate targets, one of the most coveted goals has been the prevention of virus packaging. In the cell, virus particles are made as individual entities and roam around at random until a signal is sent out for the parts to self-assemble into its infectious whole. For 30 years that signal has been known to be contained within the viral RNA. It was first found in herpes and cytomegalovirus, called the alpha sequence. Since then, other genetic elements – known as packaging signals – have been found in other viral families including coronaviruses, hepatitis viruses, and HIV.
Most of these packaging signals have been identified through indirect means as a consequence of experimental treatment. Studies have shown the necessity of these sequences but as to what exactly they do has been elusive. Experimental analyses have suggested two possible routes. The first is through a signal-based order in which an individual component binds to the signal and is prompted to package. The other is through a purely electrostatic mechanism in which the RNA acts as a spatial linchpin. It provides an environment where proteins can come together and experience a lowering of the bonding energy threshold. When this happens, the nearby proteins can thermodynamically bind to each other and start a cascade to form a mature virus.
From a purely biophysical perspective, the latter would appear to be the most viable choice. The RNA would require none of the energy required to form bonds; it would simply provide the static energy needed to facilitate protein-protein bonding. The process would be efficient and rapid.
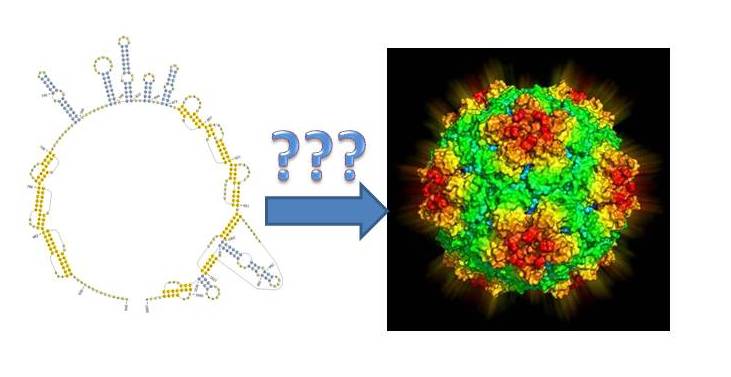
Last week, the linchpin model received a significant boost. A British team of researchers unveiled the mechanism behind electrostatic packaging. Using Satellite Tobacco Necrosis Virus, one of the simplest viruses known due to its composition of only a single coat protein and an RNA genome, the authors elucidated exactly how the process occurs. Perhaps more importantly, they also learned how this function may be conserved among numerous viruses and could help to develop therapies for the future.
The first step was to identify any actual packaging signals in the genome. This was a straightforward process in which potential RNA binding sequences within the coat protein were identified and then tested for their ability to bind to artificially made sequences. Not surprisingly, several sequences were found, supported by different secondary stem-loop structures of viral RNA.
At this point, the authors attempted to identify how the coat protein forms the actual mature virus. As expected, the coat proteins interacted with the RNA signals – bur did not bind – enabling several proteins to come into close proximity. At this point, the proteins formed larger structures around the RNA. This second step also depended on the RNA as it helped to lower the electrostatic barrier between two individual proteins.
With these two steps in the mechanism understood, the team went on to look at whether there was synergy in the function of the signals. Although the elements did not interact with each other directly, they worked co-operatively to bring coat proteins together and to fit them in the same way as a puzzle. In the presence of lower electrostatic forces, the proteins would effectively self-assemble until the entire capsid was formed.
The final stage involved a closer examination of the nature of the coat protein sequence and whether it had similarities to other viruses. The answer came with the observation the signal-interaction region contained several arginine-rich motifs. These motifs are common among many RNA viruses as they aid in the interaction and binding of RNA.
The results suggest this simplistic virus may provide a blueprint for much more complex viral entities. By looking for these packaging signals, researchers may be able to find the linchpins for virus formation. This could not only help to identify new RNAi targets for specific pathogens but, if these are evolutionarily conserved regions, may offer a path towards broader spectrum treatments and possibly vaccines.