
In 2003, scientists with the Human Genome Project announced the completion of their 13-year effort to identify the three billion base pairs that form the chemical rungs in DNA’s signature twisted-ladder shape. This first attempt to create a comprehensive map of a human biological system was more than just a breakthrough for geneticists, though. It also marked the launch of a new era of “-omic”-based research, in which biologists began shifting their attention from the individual parts within a system to studying the system itself.
“The Human Genome Project has given us a foundational change in the way we think about human biology,” says Michael Huerta, the project director for the Human Connectome Project, an effort to map neural networks in the brain. “If we have this fundamental data type, a comprehensive, systematically collected data set, it could serve as the intellectual platform for all kinds of new research directions.”
Now researchers in a variety of fields are attempting to create -omes of their own. Some of them, such as the “unknome,” the collection of all genes of unknown function, have passed quickly into obscurity. But the field now has its own journal—OMICS: A Journal of Integrative Biology—and at least four major systems have joined the ranks of the genome as subjects fit for their own major research projects.
Transcriptome
Goal: Catalog all the RNA transcripts in the body
Knowing the DNA sequence within the genome was just the first step in understanding how cells work. DNA provides the instructions (genes) for making the proteins that do most of the work of the body. But a given cell must copy these instructions before it can use them. These copies, or “transcripts,” of the DNA information are made of RNA.
In 2003, the National Institutes of Health launched the Human Transcriptome Project to catalog the transcripts. But there’s a catch: Depending on how gene transcripts are processed by the cell, our 20,000 known genes can generate hundreds of thousands of possible RNA transcripts. And it turns out that only some of these transcripts code for proteins. Others are not involved with protein synthesis; instead, these can actually affect gene expression. Still other transcripts have no known function.
Thomas R. Gingeras, a molecular biologist at Cold Spring Harbor Laboratory and the principal investigator for the project, says that the endpoint is uncertain. But already they have begun to make useful findings —for instance, their work has led to the identification of a new class of RNA transcript, called lincRNA, that is vital to cell function and could be used to fight cancer.
Proteome
Goal: Characterize all the proteins made by human cells
Proteins are the workhorses of all cells in the human body. They do everything from transporting oxygen through the bloodstream to regulating digestion. They also form the cellular structure unique to each organ or tissue type. No one yet knows how many different forms of protein there are, though, or what they all do. As mentioned, our 20,000 known genes produce multiple RNA transcripts, which can also create multiple proteins.
Moreover, proteins constantly change shape and can be chemically modified by other proteins to perform specific tasks within the cell. The result is that the genome can produce perhaps as many as a million unique proteins. The Human Proteome Project, an international collaboration formed last September, is not even close to identifying most of them. But by 2015, it aims to identify at least one protein made from each of the 20,000 protein-coding genes.
Unlike the relatively constant genome, the proteome is a dynamic network. Understanding it, says Gilbert S. Omenn, the director of the Center for Computational Medicine and Bioinformatics at the University of Michigan and a leader on the project, “is a forever activity.”
Metabolome
Goal: Trace every chemical generated by cell metabolism
The by-products our cells create as the result of their everyday functioning are called metabolites. The Human Metabolome Project was the first comprehensive attempt at characterizing all of them. The project, completed in 2008, links 3,000 metabolites to about 100 data fields, including tissue concentrations, associated diseases, and related proteins and genes. But project director David Wishart, a professor of biology and computer science from the University of Alberta, and his colleagues continue to “fill in the holes.”
Today they have identified 8,500 metabolites found in nearly every person, as well as 10,000 types that could come from external sources, such as toxins. Because metabolites are continuously generated, they are superb indicators of health; clinical chemists have used them for more than a century in diagnostics of blood and urine. Already, the metabolome project findings have provided biomarkers for hard-to-diagnose diseases like rheumatoid arthritis.
A single mutation in an enzyme can alter its efficiency, which can lead to a 10,000-fold change in the rate at which it produces metabolites. This kind of change is far easier to detect in a lab test than a gene mutation.
“Metabolites,” Wishart says, “are the canaries in the genome.”
Connectome
Goal: Build a map of the neural circuitry in the human brain
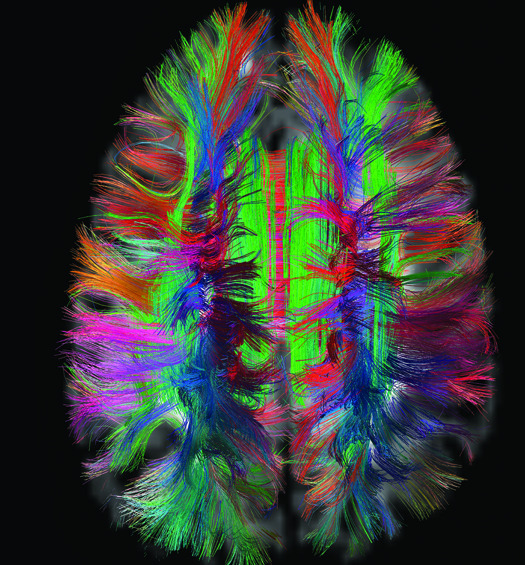
The human brain is made up of 100 billion interconnected neurons that determine our every thought and motion. Brain specialists have long gathered information about individual neurons, but no single road map exists for the entire system. The Human Connectome Project is the first major attempt to remedy that. Since 2009, researchers have been collecting functional and structural connectome data from 1,200 healthy adults.
Once complete, the project data set will provide a baseline for additional studies on the brains of people from different age groups or with diseases or disorders such as schizophrenia or alcoholism.
To create this comprehensive map of the brain, project scientists look at two types of information, brain activity and brain structure. David Van Essen of Washington University and Kamil Ugurbil of the University of Minnesota use functional MRI to determine brain activity at rest and during tasks. And to reveal the brain’s structure, the scientists use diffusion-spectrum MRI, invented by Massachusetts General Hospital neuroscientist Van Wedeen, who also works on the Connectome Project.
The project, says its director, Michael Huerta of the National Institute of Mental Health, will allow the field to “systematically ask the question, How does the brain get wired and miswired in these various disorders, as well as across a life span?”
