Gamers Solve AIDS-Related Enzyme Puzzle, Helping Scientists Search for Novel Drugs
We may earn revenue from the products available on this page and participate in affiliate programs. Learn more › Using...
We may earn revenue from the products available on this page and participate in affiliate programs. Learn more ›
Using software at home, gamers took just three weeks to solve a decade-old enzymatic enigma, in a breakthrough that could have implications for AIDS treatment. The crowdsourced puzzle solution will give scientists new insights in designing antiretroviral drugs, which could stop the AIDS virus from spreading.
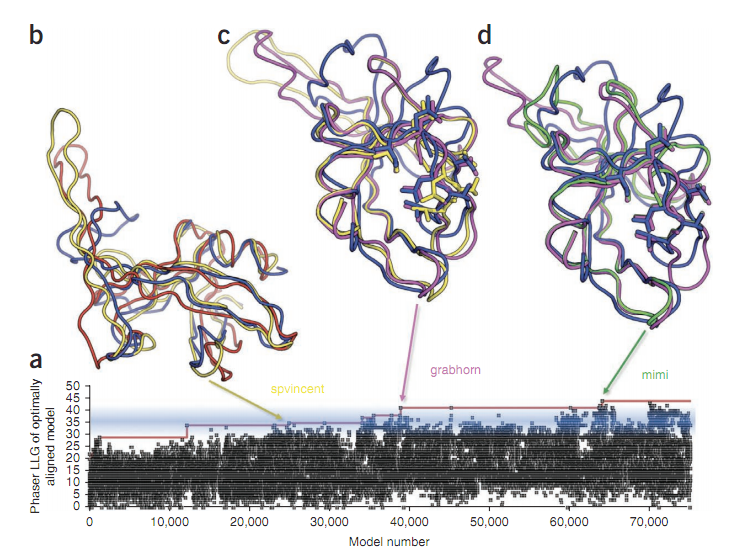
The players were able to reproduce the structure of a protein-cutting enzyme from a virus that causes an AIDS-like disease in rhesus monkeys. The enzyme, a retroviral protease, is related to how the virus spreads. Several drug studies are working on ways to block this enzyme in HIV, but this is hard to do when you don’t know what it looks like.
After about three weeks of work, the gamers’ solutions were so refined that the researchers were able to solve the entire puzzle within a couple of days. The molecular structure was so clear, the researchers could even detect surfaces where drugs might be able to attach and deactivate the enzyme. The work is described in the journal Nature Structural & Molecular Biology, which gives credit to two Foldit teams.
As we’ve described before, the University of Washington’s Foldit program lets puzzle-lovers solve complex protein-folding problems online. Scientists know the amino acid chains that make up proteins, but the ways in which these chains fold and curl into three-dimensional shapes is not well understood. Deciphering those structures can help researchers develop new drugs and therapies for cancer, Alzheimer’s, AIDS and other disorders. Foldit lets gamers give it a try, folding and twisting unique structures in ways that remain too complex for computers.
“People have spatial reasoning skills, something computers are not yet good at,” said Seth Cooper, co-creator of Foldit and a researcher at UW Department of Computing Science and Engineering, in a statement. “Games provide a framework for bringing together the strengths of computers and humans.”
The Foldit players built on each other’s work, coming up with new designs based on folds that worked and folds that failed. Over at Cosmic Log, Alan Boyle talked to one of the gamers — nicknamed “mimi” — to find out how she did it. The Nature paper’s authors said as far as they know, this is the first time gamers solved a longstanding scientific problem.
“The critical role of Foldit players in the solution of the (enzyme) structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems,” the authors write.
The best part about this news may be that there’s more to come. The UW team said they have two more papers in the pipeline, one regarding the algorithms in Foldit recipes and one regarding a brand-new synthetic protein, which was discovered through Foldit designs. Other teams may be able to tap these gamers’ creativity, leading to who knows what kind of new treatments and cures.
[via Cosmic Log]