Genetic Sequencing Gets Boost From A New Proofreading Enzyme
Error prone 3-billion-year-old process fixed
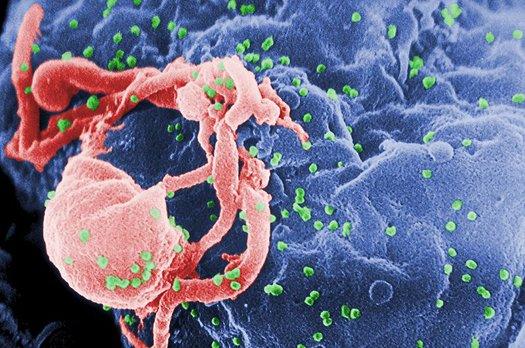
When a virus invades a cell, it uses that cell’s machinery to replicate itself by throwing the typical process for replicating genetic material into reverse. This strategy is called reverse transcription, and it’s messy, leading to numerous errors in the copied sequences. According to experts, this sloppiness might be a fluke, but it has likely turned into an evolutionary strategy to promote viral diversity and mutation, helping viruses stay ahead of the human immune response.
Since the reverse transcription process was discovered in the 70s, it has been a boon for molecular biology, allowing scientists to clone, sequence, and gain a deeper understanding of RNA. But the inherent glitchiness of the process has remained problematic.
Now a team of researchers from the University of Texas at Austin has taken the unreliability out of reverse transcription. They started with reverse transcriptase (RT), the enzyme that drives the process, and targeted a 3 billion-year-old genetic error that inhibited them from “proofreading” their gene-copying work. The new enzyme they created, called RTX, can correct its mistakes, creating far more accurate genomic sequences than ever before.
According to their paper, published in Science, RTX can produce transcriptions that are three to 10 times more accurate than what was possible before.
From the press release: