How To Watch An Animal Develop Cell By Cell
I remember when you were just two cells big!
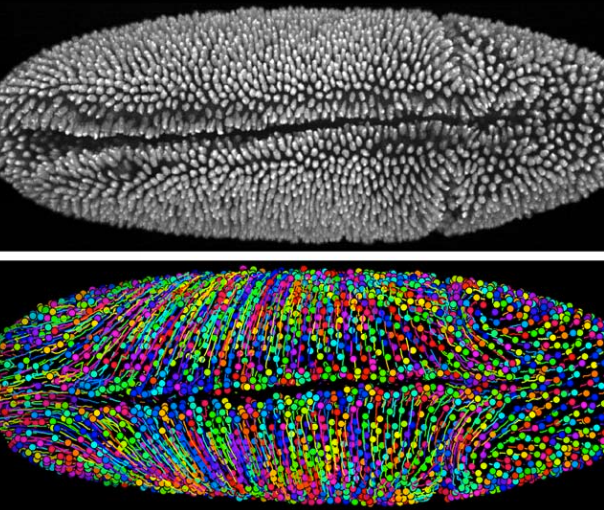
What is this fuzzy creature? Sadly, it’s not pettable. This is a microscope image of a fruit fly embryo, showing the individual cells within it. That’s 2,458 cells, to be exact. The bottom image shows each cell in a different color, with lines to show how those cells moved around.
The image comes from a new technique scientists developed to watch every single cell in an early embryo and record what it does. Where does each cell go? When does it divide into two cells, and where do each of those two cells go? How do genes control what cells do? These are some of the most basic questions in biology.
Of course, previous generations of scientists have developed their own cool methods for answering these questions. But none of the methods is perfect. A human couldn’t do such tracking without computer help, but even for a computer, it’s difficult to keep tabs on thousands of moving, duplicating cells at once. In this latest work, a team of biologists from the Howard Hughes Medical Institute is releasing, for free, its method for recording embryotic development automatically. The technique is able to track up to 20,000 cells at once and is 97 percent accurate. The team has used it to watch the development of fruit fly, zebrafish and mouse embryos—basically, how these animals are built from the ground up. The idea is that animal species are similar enough that scientists are able to learn some basic principles about how humans develop by watching lab animals, using techniques it would be unethical to apply to a human embryo.
The Howard Hughes scientists used several clever solutions to deal with the terabytes of data embryo-tracking creates. They packaged individual pixels up into “superpixels,” here called “supervoxels” because they’re 3-D. The biologists’ computer program also automatically recognizes and fixes parts of its dataset where it might have made mistakes in tracking. (Like human eyes, computer vision can have trouble distinguishing cells if they’re too close together, which often happens in embryos.) The result is a program that’s more accurate and more than 10 times faster than previous programs, according to a paper the team published this week in the journal Nature Methods.